You are here : FHU-SEPSISENOBJECTIVES & PROJECTSRoad MapWP3 – Multi-omics platform
- Partager cette page :
- PDF version
WP3 – Multi-omics platform
Leader : Pr Stanislas Grassin-Delyle |
Co-leader : Pr Henri-Jean Garchon |
RATIONAL AND OBJECTIVE
The objective of the Sepsis FHU is to develop "point of care" tests as an aid to the rapid diagnosis of patients with sepsis and as a guide to the personalizing hemodynamic, renal, metabolic and immune interventions. This requires our teams to identify biomarkers of interest using the most efficient technologies. Over the past decade, omics technologies have gained increasing traction. Integrated multi-omic approaches combine the interests and specificities of genomics, transcriptomics and metabolomics. Genomic and transcriptomic approaches enable an in-depth analysis of genetic variation and gene expression. Metabolomics is devoted to the study of metabolites, small molecular weight molecules, mainly by mass spectrometry. These approaches are usually implemented in biological fluids or tissues, but several hundred volatile metabolites, called volatile organic compounds, can also be measured in exhaled breath where they reflect the metabolic activity of the organism and pathology.
Several platforms and laboratories within the UVSQ/Université Paris-Saclay will be involved in the FHU's research projects. Firstly, the platforms of the Department of Health Biotechnology (UFR Simone Veil - Santé), including the genomic and mass spectrometry platforms, which have the instruments and expertise necessary to implement all the "omics" approaches. The CNRGH platform (CEA) will also be called upon for whole genome sequencing. Secondly, the Exhalomics® platform (Hôpital Foch/UVSQ) has a range of technological tools such as electronic noses and mass spectrometers dedicated to clinical research projects for exhaled breath analysis. Data processing will be partly carried out by the platforms concerned, but also in collaboration with the Data Science for Molecular Phenotyping and Precision Medicine team (SciDoPhenIA, Dr Thévenot, CEA, Department of Drug Technology and Life Sciences), which specializes in the processing, analysis and integration of metabolomics and volatolomics data.
The strength of the FHU will be to offer these platforms perfectly characterized large cohort samples of patients, based on previously established cohorts and the RHU RECORDS adaptive clinical trial.
ORGANIZATION
The general organization of the WP is shown in Figure 1 below. Integrated multi-omic signatures will be established from existing or ongoing cohorts, using technological tools made available by a set of platforms and using state-of-the-art statistical methods that integrate high-level multidimensional dataset generated. Established signatures will then be forwarded to WP2 for integration with the higher level, including all demographic, clinical, biological, therapeutic and epidemiological data.
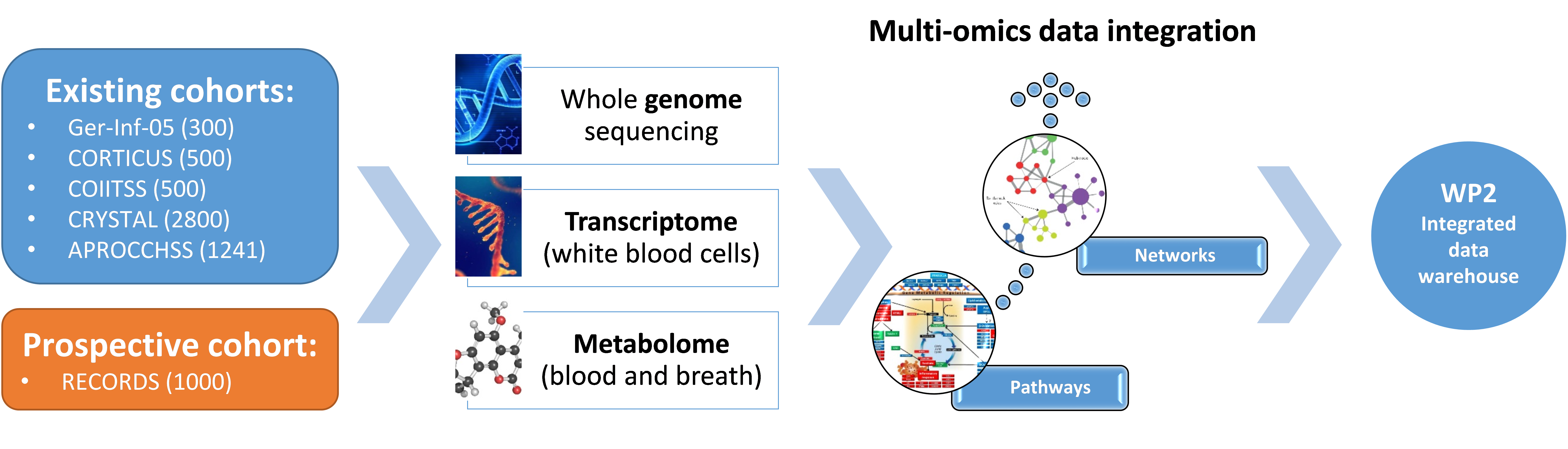
Figure 1: Objectives and organization of WP3

